Replisome Mediated DNA replication. During DNA replication, the double-helical DNA needs to be separated for replication 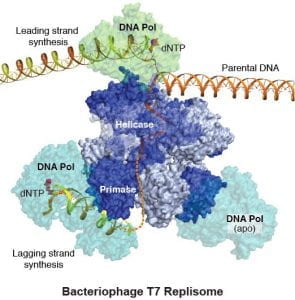 by a helicase motor. Each strand is then copied by DNA polymerases, continuously on the leading strand and discontinuously (via Okazaki fragments) on the lagging strand, where each DNA synthesis initiates from an RNA primer provided by primase. The helicase, polymerase, and primase assemble at a replication fork as an integer complex named “replisome” and cooperatively perform the DNA replication. Other factors, such as nucleases, single-strand binding proteins, processivity factors for DNA polymerases, and adaptors coordinates helicase with polymerase or primases, and enhances the performance of replisomes. Although equipped with same enzymatic activities, replisomes from different kingdoms of life are not evolutionarily conserved and with varied complexity. We are investigating the molecular mechanisms of replisome operation. Our previous work on Bacteriophage T7 replisome offers the first glimpse of how helicase, primase and polymerases are organized at a DNA replication fork and how different activities are coordinated.
by a helicase motor. Each strand is then copied by DNA polymerases, continuously on the leading strand and discontinuously (via Okazaki fragments) on the lagging strand, where each DNA synthesis initiates from an RNA primer provided by primase. The helicase, polymerase, and primase assemble at a replication fork as an integer complex named “replisome” and cooperatively perform the DNA replication. Other factors, such as nucleases, single-strand binding proteins, processivity factors for DNA polymerases, and adaptors coordinates helicase with polymerase or primases, and enhances the performance of replisomes. Although equipped with same enzymatic activities, replisomes from different kingdoms of life are not evolutionarily conserved and with varied complexity. We are investigating the molecular mechanisms of replisome operation. Our previous work on Bacteriophage T7 replisome offers the first glimpse of how helicase, primase and polymerases are organized at a DNA replication fork and how different activities are coordinated.
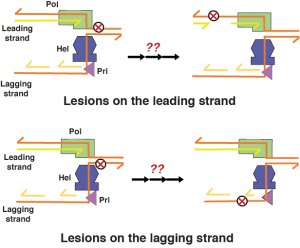 Replisome Stress Response. The genomic DNA are susceptible to various endogenous and environment assaults. Damages on DNA can slow down or block replication progression and induce replication stress response. Enzymes within a replisome will be the first to hit any DNA roadblock. How a replisome reorganizes in response to different roadblocks and signals for replication stress remain elusive. Moreover, replisome has evolved amazing abilities to circumvent DNA roadblocks and reassume DNA synthesis downstream, leaving the lesions repaired post-replicationally. With the simple model replisome from Bacteriophage T7, we hope to uncover the structural basis of how a replisome response to and traverse different roadblocks on DNA.
Replisome Stress Response. The genomic DNA are susceptible to various endogenous and environment assaults. Damages on DNA can slow down or block replication progression and induce replication stress response. Enzymes within a replisome will be the first to hit any DNA roadblock. How a replisome reorganizes in response to different roadblocks and signals for replication stress remain elusive. Moreover, replisome has evolved amazing abilities to circumvent DNA roadblocks and reassume DNA synthesis downstream, leaving the lesions repaired post-replicationally. With the simple model replisome from Bacteriophage T7, we hope to uncover the structural basis of how a replisome response to and traverse different roadblocks on DNA.
Helicase Translocation. Helicases are chemo-mechanical motors that couple the energy from ATP hydrolysis to 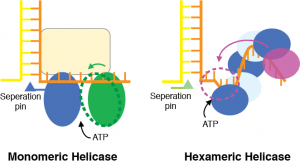 DNA translocation. They can separate double-stranded DNA or RNA, anneal single strands to a duplex, remove proteins on DNA or RNA, and remodel replication fork, Holliday junction and chromatin structures. Corresponding to their diverse functions, helicases are with different substrate preference, mechanistic properties and molecular organizations. We studies helicases in DNA replication and replication stress response. We would like to learn the structural basis of their energy coupling, translocation, and regulation.
DNA translocation. They can separate double-stranded DNA or RNA, anneal single strands to a duplex, remove proteins on DNA or RNA, and remodel replication fork, Holliday junction and chromatin structures. Corresponding to their diverse functions, helicases are with different substrate preference, mechanistic properties and molecular organizations. We studies helicases in DNA replication and replication stress response. We would like to learn the structural basis of their energy coupling, translocation, and regulation.
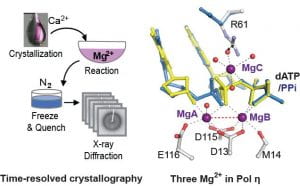 Mg2+-Dependent DNA Synthesis and Degradation. Magnesium ions (Mg2+) are essential co-factors for many enzymes in nucleic acid metabolism. Mg2+ can bridge negatively charged substrates, stabilize transition states, and promote hydroxyl group deprotonation and nucleophilic attack. For decades, two Mg2+ ions have been proposed to be required and sufficient for the catalysis of polymerases and many nucleases. We have observed the human DNA polymerase η catalysis with time-resolved crystallography. Surprisingly, we found the capture of a transiently bound third Mg2+ ion during the chemical reaction. Similarly, transiently bound Mg2+ ion was subsequently reported in other polymerases and nucleases that were thought to be two-Mg2+-ion-dependent. We are further exploring the role of the transiently bound Mg2+ during polymerase catalysis, nucleotide misincorporation and nucleoside-analog drug inhibition.
Mg2+-Dependent DNA Synthesis and Degradation. Magnesium ions (Mg2+) are essential co-factors for many enzymes in nucleic acid metabolism. Mg2+ can bridge negatively charged substrates, stabilize transition states, and promote hydroxyl group deprotonation and nucleophilic attack. For decades, two Mg2+ ions have been proposed to be required and sufficient for the catalysis of polymerases and many nucleases. We have observed the human DNA polymerase η catalysis with time-resolved crystallography. Surprisingly, we found the capture of a transiently bound third Mg2+ ion during the chemical reaction. Similarly, transiently bound Mg2+ ion was subsequently reported in other polymerases and nucleases that were thought to be two-Mg2+-ion-dependent. We are further exploring the role of the transiently bound Mg2+ during polymerase catalysis, nucleotide misincorporation and nucleoside-analog drug inhibition.